| Gene Symbol | ATP5IF1 |
| Entrez Gene ID | 327699 |
| Full Name | ATP synthase inhibitory factor subunit 1 |
| Synonyms | ATPIF1 |
| Gene Type | protein-coding |
| Organism | Bos taurus(cattle) |
HOME
ORF » Species Summary » Bos taurus » ATP5IF1 cDNA ORF clone
| Gene Symbol | ATP5IF1 |
| Entrez Gene ID | 327699 |
| Full Name | ATP synthase inhibitory factor subunit 1 |
| Synonyms | ATPIF1 |
| Gene Type | protein-coding |
| Organism | Bos taurus(cattle) |
| mRNA | Protein | Name |
|---|---|---|
| NM_175816.3 | NP_787010.1 | ATPase inhibitor, mitochondrial precursor |

| Pan troglodytes (chimpanzee) | ATPIF1 | XP_003339032.1 |
| Danio rerio (zebrafish) | atpif1a | NP_001082990.1 |
| Rattus norvegicus (Norway rat) | Atpif1 | NP_037047.2 |
| ATPIF1 | XP_001113002.1 | |
| Canis lupus familiaris (dog) | ATPIF1 | XP_865710.1 |
| Homo sapiens (human) | ATPIF1 | NP_057395.1 |
| Bos taurus (cattle) | ATPIF1 | NP_787010.1 |
| Mus musculus (house mouse) | Atpif1 | NP_031538.2 |
| Danio rerio (zebrafish) | atpif1b | NP_001038324.1 |
| Xenopus tropicalis (tropical clawed frog) | atpif1 | NP_001135565.1 |

Pathway of binding of the intrinsically disordered mitochondrial inhibitor protein to F1-ATPase.
Bason JV, Montgomery MG, Leslie AG, Walker JE
Proceedings of the National Academy of Sciences of the United States of America111(31)11305-10(2014 Aug)
Binding of the inhibitor protein IF(1) to bovine F(1)-ATPase.
Bason JV, Runswick MJ, Fearnley IM, Walker JE
Journal of molecular biology406(3)443-53(2011 Feb)
A whole-genome assembly of the domestic cow, Bos taurus.
Zimin AV, Delcher AL, Florea L, Kelley DR, Schatz MC, Puiu D, Hanrahan F, Pertea G, Van Tassell CP, Sonstegard TS, Mar?ais G, Roberts M, Subramanian P, Yorke JA, Salzberg SL
Genome biology10(4)R42(2009)
Glutamic acid in the inhibitory site of mitochondrial ATPase inhibitor, IF(1), participates in pH sensing in both mammals and yeast.
Ando C, Ichikawa N
Journal of biochemistry144(4)547-53(2008 Oct)
Identification of a conserved calmodulin-binding motif in the sequence of F0F1 ATPsynthase inhibitor protein.
Contessi S, Haraux F, Mavelli I, Lippe G
Journal of bioenergetics and biomembranes37(5)317-26(2005 Oct)
GeneRIFs: Gene References Into Functions What's a GeneRIF?
The intrinsically disordered inhibitory region of IF1 interacts first with the alphaEbetaE-catalytic interface, the most open of the three catalytic interfaces, where the available interactions with the enzyme allow it to form an alpha-helix from residues 31-49.
Title: Pathway of binding of the intrinsically disordered mitochondrial inhibitor protein to F1-ATPase.
The N-terminal region of IF(1) destabilizes the interaction of the inhibitor with F(1)-ATPase and may assist in removing the inhibitor from its binding site when F(1)F(o)-ATPase is making ATP
Title: Binding of the inhibitor protein IF(1) to bovine F(1)-ATPase.
in addition to His49, Glu26 participates in pH sensing in bovine IF(1), and the mechanism of pH sensing mediated by Glu26 is different from the dimer-tetramer model proposed previously
Title: Glutamic acid in the inhibitory site of mitochondrial ATPase inhibitor, IF(1), participates in pH sensing in both mammals and yeast.
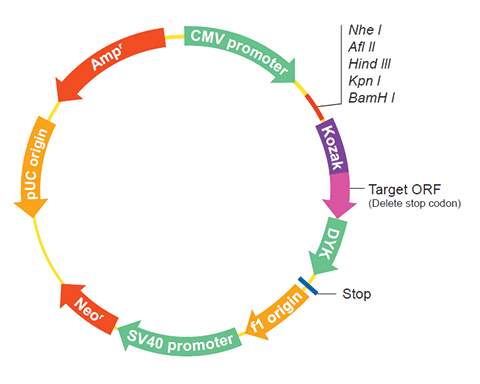
The following ATP5IF1 gene cDNA ORF clone sequences were retrieved from the NCBI Reference Sequence Database (RefSeq). These sequences represent the protein coding region of the ATP5IF1 cDNA ORF which is encoded by the open reading frame (ORF) sequence. ORF sequences can be delivered in our standard vector, pcDNA3.1+/C-(K)DYK or the vector of your choice as an expression/transfection-ready ORF clone. Not the clone you want? Click here to find your clone.
| CloneID | OBa01301 | |
| Clone ID Related Accession (Same CDS sequence) | NM_175816.3 | |
| Accession Version | NM_175816.3 Latest version! | Documents for ORF clone product in default vector |
| Sequence Information | ORF Nucleotide Sequence (Length: 330bp) Protein sequence SNP |
|
| Vector | pcDNA3.1-C-(k)DYK or customized vector |  User Manual User Manual |
| Clone information | Clone Map |  MSDS MSDS |
| Tag on pcDNA3.1+/C-(K)DYK | C terminal DYKDDDDK tags | |
| ORF Insert Method | CloneEZ™ Seamless cloning technology | |
| Insert Structure | linear | |
| Update Date | 2019-12-25 | |
| Organism | Bos taurus(cattle) | |
| Product | ATPase inhibitor, mitochondrial precursor | |
| Comment | Comment: PROVISIONAL REFSEQ: This record has not yet been subject to final NCBI review. The reference sequence was derived from BC111645.1. On Aug 30, 2012 this sequence version replaced NM_175816.2. Publication Note: This RefSeq record includes a subset of the publications that are available for this gene. Please see the Gene record to access additional publications. ##Evidence-Data-START## Transcript exon combination :: BC111645.1, DV906348.1 [ECO:0000332] RNAseq introns :: single sample supports all introns SAMN03145413, SAMN03145415 [ECO:0000348] ##Evidence-Data-END## ##RefSeq-Attributes-START## gene product(s) localized to mito. :: inferred from homology ##RefSeq-Attributes-END## | |
1 | ATGGCAGCCA CGGCGTTGGC GGCGCGTACC CGGCAGGCCG TCTGGAGCGT CTGGGCAATG |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
| RefSeq | NP_787010.1 |
| CDS | 6..335 |
| Translation |

Target ORF information:
Target ORF information:
|
 NM_175816.3 NM_175816.3 |

1 | ATGGCAGCCA CGGCGTTGGC GGCGCGTACC CGGCAGGCCG TCTGGAGCGT CTGGGCAATG |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
Pathway of binding of the intrinsically disordered mitochondrial inhibitor protein to F1-ATPase. |
Binding of the inhibitor protein IF(1) to bovine F(1)-ATPase. |
A whole-genome assembly of the domestic cow, Bos taurus. |
Glutamic acid in the inhibitory site of mitochondrial ATPase inhibitor, IF(1), participates in pH sensing in both mammals and yeast. |
Identification of a conserved calmodulin-binding motif in the sequence of F0F1 ATPsynthase inhibitor protein. |