| Gene Symbol | FUN30 |
| Entrez Gene ID | 851214 |
| Full Name | DNA-dependent ATPase FUN30 |
| Gene Type | protein-coding |
| Organism | Saccharomyces cerevisiae S288C |
HOME
ORF » Species Summary » Saccharomyces cerevisiae S288C » FUN30 cDNA ORF clone
| Gene Symbol | FUN30 |
| Entrez Gene ID | 851214 |
| Full Name | DNA-dependent ATPase FUN30 |
| Gene Type | protein-coding |
| Organism | Saccharomyces cerevisiae S288C |
| mRNA | Protein | Name |
|---|---|---|
| NM_001178164.1 | NP_009383.1 | DNA-dependent ATPase FUN30 |

| Schizosaccharomyces pombe (fission yeast) | fft2 | NP_587731.1 |
| Saccharomyces cerevisiae (baker's yeast) | FUN30 | NP_009383.1 |
| Kluyveromyces lactis | KLLA0F07513g | XP_455420.1 |
| Schizosaccharomyces pombe (fission yeast) | fft3 | NP_593617.1 |
| Eremothecium gossypii | AGOS_ACR286C | NP_983688.1 |
| Schizosaccharomyces pombe (fission yeast) | fft1 | NP_593325.1 |

Life with 6000 genes.
Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG
Science (New York, N.Y.)274(5287)546, 563-7(1996 Oct)
The nucleotide sequence of chromosome I from Saccharomyces cerevisiae.
Bussey H, Kaback DB, Zhong W, Vo DT, Clark MW, Fortin N, Hall J, Ouellette BF, Keng T, Barton AB
Proceedings of the National Academy of Sciences of the United States of America92(9)3809-13(1995 Apr)
GeneRIFs: Gene References Into Functions What's a GeneRIF?
Fun30 acts to down regulate Rad9-dependent DNA damage checkpoint triggered by CPT or MMS, but does not affect Rad9-independent intra-S phase replication checkpoint induced by MMS or HU. It supports the notion that Fun30 contributes to cellular response to DSBs by preventing excessive DNA damage checkpoint activation in addition to its role in facilitating DNA end resection.
Title: Cell cycle-dependent positive and negative functions of Fun30 chromatin remodeler in DNA damage response.
Budding yeast Fun30 and human SMARCAD1 are cell cycle-regulated by interaction with the DNA double strand break-localized scaffold protein Dpb11/TOPBP1, respectively. In yeast, this protein assembly additionally comprises the 9-1-1 damage sensor, is involved in localizing Fun30 to damaged chromatin, and thus is required for efficient long-range resection of DNA double strand breaks.
Title: Targeting of the Fun30 nucleosome remodeller by the Dpb11 scaffold facilitates cell cycle-regulated DNA end resection.
Cdk1 and multiple cyclins become highly enriched at DSBs and that the recruitment of Cdk1 and cyclins Clb2 and Clb5 ensures optimal Fun30 phosphorylation and checkpoint activation.
Title: Enrichment of Cdk1-cyclins at DNA double-strand breaks stimulates Fun30 phosphorylation and DNA end resection.
These results define a role for Fun30 in the regulation of transcription and indicate that Fun30 remodels chromatin at the 5' end of genes by sliding promoter-proximal nucleosomes.
Title: The ATP-dependent chromatin remodeling enzyme Fun30 represses transcription by sliding promoter-proximal nucleosomes.
Fun30, a poorly characterized member of the Swi2/Snf2 family of chromatin remodelers, plays a role in end processing by facilitating the Exo1 and Sgs1-Dna2 resection pathways.
Title: Overcoming the chromatin barrier to end resection.
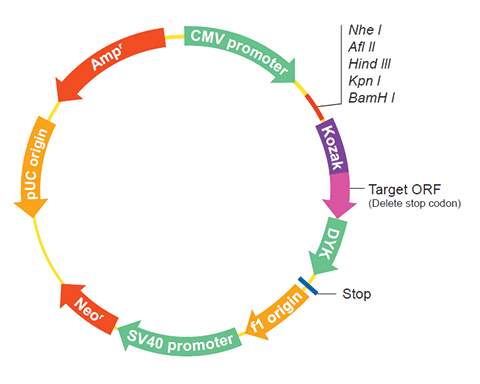
The following FUN30 gene cDNA ORF clone sequences were retrieved from the NCBI Reference Sequence Database (RefSeq). These sequences represent the protein coding region of the FUN30 cDNA ORF which is encoded by the open reading frame (ORF) sequence. ORF sequences can be delivered in our standard vector, pcDNA3.1+/C-(K)DYK or the vector of your choice as an expression/transfection-ready ORF clone. Not the clone you want? Click here to find your clone.
| CloneID | OSi00800 | |
| Clone ID Related Accession (Same CDS sequence) | NM_001178164.1 | |
| Accession Version | NM_001178164.1 Latest version! | Documents for ORF clone product in default vector |
| Sequence Information | ORF Nucleotide Sequence (Length: 3396bp) Protein sequence SNP |
|
| Vector | pcDNA3.1-C-(k)DYK or customized vector |  User Manual User Manual |
| Clone information | Clone Map |  MSDS MSDS |
| Tag on pcDNA3.1+/C-(K)DYK | C terminal DYKDDDDK tags | |
| ORF Insert Method | CloneEZ™ Seamless cloning technology | |
| Insert Structure | linear | |
| Update Date | 2019-10-31 | |
| Organism | Saccharomyces cerevisiae S288C | |
| Product | DNA-dependent ATPase FUN30 | |
| Comment | Comment: REVIEWED REFSEQ: This record has been curated by SGD. This record is derived from an annotated genomic sequence (NC_001133). ##Genome-Annotation-Data-START## Annotation Provider :: SGD Annotation Status :: Full Annotation Annotation Version :: R64-2-1 URL :: http://www.yeastgenome.org/ ##Genome-Annotation-Data-END## COMPLETENESS: incomplete on both ends. | |
1 | ATGAGTGGTT CGCATTCAAA TGATGAGGAT GACGTAGTGC AAGTGCCCGA GACGTCCTCT |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
| RefSeq | NP_009383.1 |
| CDS | 1..3396 |
| Translation |

Target ORF information:
Target ORF information:
|
 NM_001178164.1 NM_001178164.1 |

1 | ATGAGTGGTT CGCATTCAAA TGATGAGGAT GACGTAGTGC AAGTGCCCGA GACGTCCTCT |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
Life with 6000 genes. |
The nucleotide sequence of chromosome I from Saccharomyces cerevisiae. |