| Gene Symbol | HSP12 |
| Entrez Gene ID | 850532 |
| Full Name | lipid-binding protein HSP12 |
| Synonyms | GLP1,HOR5 |
| Gene Type | protein-coding |
| Organism | Saccharomyces cerevisiae S288C |
HOME
ORF » Species Summary » Saccharomyces cerevisiae S288C » HSP12 cDNA ORF clone
| Gene Symbol | HSP12 |
| Entrez Gene ID | 850532 |
| Full Name | lipid-binding protein HSP12 |
| Synonyms | GLP1,HOR5 |
| Gene Type | protein-coding |
| Organism | Saccharomyces cerevisiae S288C |
| mRNA | Protein | Name |
|---|---|---|
| NM_001179952.1 | NP_116640.1 | lipid-binding protein HSP12 |

| Saccharomyces cerevisiae (baker's yeast) | HSP12 | NP_116640.1 |
| Kluyveromyces lactis | KLLA0D07634g | XP_453403.1 |

Life with 6000 genes.
Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG
Science (New York, N.Y.)274(5287)546, 563-7(1996 Oct)
Analysis of the nucleotide sequence of chromosome VI from Saccharomyces cerevisiae.
Murakami Y, Naitou M, Hagiwara H, Shibata T, Ozawa M, Sasanuma S, Sasanuma M, Tsuchiya Y, Soeda E, Yokoyama K
Nature genetics10(3)261-8(1995 Jul)
GeneRIFs: Gene References Into Functions What's a GeneRIF?
these results demonstrate that the combined action of Hsp12p, Pau5p and a killer toxin is sufficient to steer a yeast community.
Title: Hsp12p and PAU genes are involved in ecological interactions between natural yeast strains.
Hsp12 expression contributes to lifespan extension, possibly via membrane alterations
Title: NMR structure of Hsp12, a protein induced by and required for dietary restriction-induced lifespan extension in yeast.
NMR spectra collected in presence of paramagnetic spin relaxation agents (5DSA, 16DSA, and Gd(DTPA-BMA)) indicated that the amphipathic alpha-helices of Hsp12 in SDS micelles lie on the membrane surface.
Title: Structural characterization of Hsp12, the heat shock protein from Saccharomyces cerevisiae, in aqueous solution where it is intrinsically disordered and in detergent micelles where it is locally ?-helical.
Hsp12 is completely unfolded; however, in the presence of certain lipids, it adopts a helical structure. The presence of Hsp12 does not alter the overall lipid composition of the plasma membrane but increases membrane stability.
Title: Hsp12 is an intrinsically unstructured stress protein that folds upon membrane association and modulates membrane function.
The decrease of the turgor pressure in the case of WT strain upon addition of Congo red is shown to be consistent with an upregulation of Hsp12p in the close vicinity of the plasma membrane.
Title: The role of the heat shock protein Hsp12p in the dynamic response of Saccharomyces cerevisiae to the addition of Congo red.
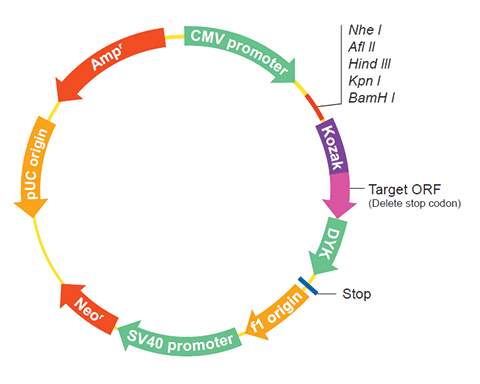
The following HSP12 gene cDNA ORF clone sequences were retrieved from the NCBI Reference Sequence Database (RefSeq). These sequences represent the protein coding region of the HSP12 cDNA ORF which is encoded by the open reading frame (ORF) sequence. ORF sequences can be delivered in our standard vector, pcDNA3.1+/C-(K)DYK or the vector of your choice as an expression/transfection-ready ORF clone. Not the clone you want? Click here to find your clone.
| CloneID | OSi00221 | |
| Clone ID Related Accession (Same CDS sequence) | NM_001179952.1 | |
| Accession Version | NM_001179952.1 Latest version! | Documents for ORF clone product in default vector |
| Sequence Information | ORF Nucleotide Sequence (Length: 330bp) Protein sequence SNP |
|
| Vector | pcDNA3.1-C-(k)DYK or customized vector |  User Manual User Manual |
| Clone information | Clone Map |  MSDS MSDS |
| Tag on pcDNA3.1+/C-(K)DYK | C terminal DYKDDDDK tags | |
| ORF Insert Method | CloneEZ™ Seamless cloning technology | |
| Insert Structure | linear | |
| Update Date | 2019-10-31 | |
| Organism | Saccharomyces cerevisiae S288C | |
| Product | lipid-binding protein HSP12 | |
| Comment | Comment: REVIEWED REFSEQ: This record has been curated by SGD. This record is derived from an annotated genomic sequence (NC_001138). ##Genome-Annotation-Data-START## Annotation Provider :: SGD Annotation Status :: Full Annotation Annotation Version :: R64-2-1 URL :: http://www.yeastgenome.org/ ##Genome-Annotation-Data-END## COMPLETENESS: incomplete on both ends. | |
1 | ATGTCTGACG CAGGTAGAAA AGGATTCGGT GAAAAAGCTT CTGAAGCTTT GAAGCCAGAC |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
| RefSeq | NP_116640.1 |
| CDS | 1..330 |
| Translation |

Target ORF information:
Target ORF information:
|
 NM_001179952.1 NM_001179952.1 |

1 | ATGTCTGACG CAGGTAGAAA AGGATTCGGT GAAAAAGCTT CTGAAGCTTT GAAGCCAGAC |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
Life with 6000 genes. |
Analysis of the nucleotide sequence of chromosome VI from Saccharomyces cerevisiae. |