| Gene Symbol | RNR1 |
| Entrez Gene ID | 856801 |
| Full Name | ribonucleotide-diphosphate reductase subunit RNR1 |
| Synonyms | CRT7,RIR1,SDS12 |
| Gene Type | protein-coding |
| Organism | Saccharomyces cerevisiae S288C |
HOME
ORF » Species Summary » Saccharomyces cerevisiae S288C » RNR1 cDNA ORF clone
| Gene Symbol | RNR1 |
| Entrez Gene ID | 856801 |
| Full Name | ribonucleotide-diphosphate reductase subunit RNR1 |
| Synonyms | CRT7,RIR1,SDS12 |
| Gene Type | protein-coding |
| Organism | Saccharomyces cerevisiae S288C |
| mRNA | Protein | Name |
|---|---|---|
| NM_001178961.1 | NP_010993.1 | ribonucleotide-diphosphate reductase subunit RNR1 |

| Canis lupus familiaris (dog) | RRM1 | XP_534027.2 |
| Mus musculus (house mouse) | Rrm1 | NP_033129.2 |
| Gallus gallus (chicken) | RRM1 | NP_001026008.1 |
| Saccharomyces cerevisiae (baker's yeast) | RNR3 | NP_012198.3 |
| Eremothecium gossypii | AGOS_ADL057W | NP_984039.2 |
| Schizosaccharomyces pombe (fission yeast) | cdc22 | NP_594491.1 |
| Pan troglodytes (chimpanzee) | RRM1 | XP_001160029.1 |
| Bos taurus (cattle) | RRM1 | NP_001106767.1 |
| Rattus norvegicus (Norway rat) | Rrm1 | NP_001013254.1 |
| Caenorhabditis elegans (roundworm) | rnr-1 | NP_499039.1 |
| Saccharomyces cerevisiae (baker's yeast) | RNR1 | NP_010993.1 |
| Oryza sativa (rice) | Os06g0168600 | NP_001056926.1 |
| Danio rerio (zebrafish) | rrm1 | NP_571530.1 |
| Magnaporthe oryzae (rice blast fungus) | MGG_07000 | XP_003709756.1 |
| Neurospora crassa | NCU03539 | XP_956818.1 |
| Xenopus tropicalis (tropical clawed frog) | rrm1 | NP_001120566.1 |
| Homo sapiens (human) | RRM1 | NP_001024.1 |
| RRM1 | XP_001113010.1 | |
| Drosophila melanogaster (fruit fly) | RnrL | NP_477027.1 |
| Anopheles gambiae (African malaria mosquito) | AgaP_AGAP010198 | XP_554103.3 |
| Kluyveromyces lactis | KLLA0C07887g | XP_452551.1 |
| Arabidopsis thaliana (thale cress) | RNR1 | NP_179770.1 |

The nucleotide sequence of Saccharomyces cerevisiae chromosome V.
Dietrich FS, Mulligan J, Hennessy K, Yelton MA, Allen E, Araujo R, Aviles E, Berno A, Brennan T, Carpenter J, Chen E, Cherry JM, Chung E, Duncan M, Guzman E, Hartzell G, Hunicke-Smith S, Hyman RW, Kayser A, Komp C, Lashkari D, Lew H, Lin D, Mosedale D, Davis RW
Nature387(6632 Suppl)78-81(1997 May)
Life with 6000 genes.
Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG
Science (New York, N.Y.)274(5287)546, 563-7(1996 Oct)
GeneRIFs: Gene References Into Functions What's a GeneRIF?
Fhl1p regulates RNR1 gene transcription to maintain dNTP levels, thus modulating longevity by protection against replication stress
Title: The forkhead-like transcription factor (Fhl1p) maintains yeast replicative lifespan by regulating ribonucleotide reductase 1 (RNR1) gene transcription.
Multiple turnover kinetics show that Sml1 inhibition of dGTP/ADP- and ATP/CDP-bound ScRR follows a mixed inhibition mechanism. However, Sml1 cooperatively binds to the ES complex in the dGTP/ADP form, whereas with ATP/CDP, Sml1 binds weakly and noncooperatively
Title: Inhibition of yeast ribonucleotide reductase by Sml1 depends on the allosteric state of the enzyme.
there is a directed orientation of the RNR beta and beta' C-terminal tails relative to alpha within the holoenzyme consistent with a docking model of the two subunits and argue against RT across the beta beta' interface
Title: Investigation of in vivo roles of the C-terminal tails of the small subunit (??') of Saccharomyces cerevisiae ribonucleotide reductase: contribution to cofactor formation and intersubunit association within the active holoenzyme.
The data reveaedl that the arginine 293 and glutamine 288 residues of ribonucleotide reductase 1 are crucial in facilitating ADP and CDP substrate selection.
Title: Role of arginine 293 and glutamine 288 in communication between catalytic and allosteric sites in yeast ribonucleotide reductase.
defects in autophagy result in an increase in soluble Rnr1 protein levels and a DNA damage phenotype.
Title: Autophagy-dependent regulation of the DNA damage response protein ribonucleotide reductase 1.
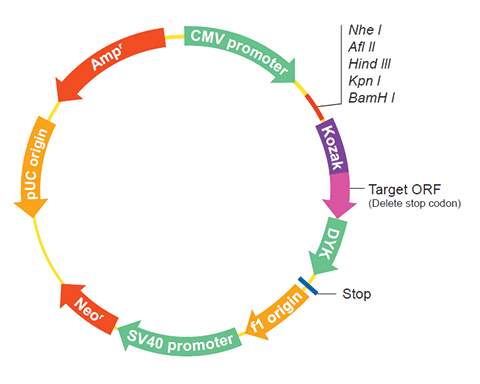
The following RNR1 gene cDNA ORF clone sequences were retrieved from the NCBI Reference Sequence Database (RefSeq). These sequences represent the protein coding region of the RNR1 cDNA ORF which is encoded by the open reading frame (ORF) sequence. ORF sequences can be delivered in our standard vector, pcDNA3.1+/C-(K)DYK or the vector of your choice as an expression/transfection-ready ORF clone. Not the clone you want? Click here to find your clone.
| CloneID | OSi05565 | |
| Clone ID Related Accession (Same CDS sequence) | NM_001178961.1 | |
| Accession Version | NM_001178961.1 Latest version! | Documents for ORF clone product in default vector |
| Sequence Information | ORF Nucleotide Sequence (Length: 2667bp) Protein sequence SNP |
|
| Vector | pcDNA3.1-C-(k)DYK or customized vector |  User Manual User Manual |
| Clone information | Clone Map |  MSDS MSDS |
| Tag on pcDNA3.1+/C-(K)DYK | C terminal DYKDDDDK tags | |
| ORF Insert Method | CloneEZ™ Seamless cloning technology | |
| Insert Structure | linear | |
| Update Date | 2019-10-31 | |
| Organism | Saccharomyces cerevisiae S288C | |
| Product | ribonucleotide-diphosphate reductase subunit RNR1 | |
| Comment | Comment: REVIEWED REFSEQ: This record has been curated by SGD. This record is derived from an annotated genomic sequence (NC_001137). ##Genome-Annotation-Data-START## Annotation Provider :: SGD Annotation Status :: Full Annotation Annotation Version :: R64-2-1 URL :: http://www.yeastgenome.org/ ##Genome-Annotation-Data-END## COMPLETENESS: incomplete on both ends. | |
1 | ATGTACGTTT ATAAAAGAGA CGGTCGTAAA GAACCTGTCC AATTCGATAA GATTACCGCT |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
| RefSeq | NP_010993.1 |
| CDS | 1..2667 |
| Translation |

Target ORF information:
Target ORF information:
|
 NM_001178961.1 NM_001178961.1 |

1 | ATGTACGTTT ATAAAAGAGA CGGTCGTAAA GAACCTGTCC AATTCGATAA GATTACCGCT |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
The nucleotide sequence of Saccharomyces cerevisiae chromosome V. |
Life with 6000 genes. |