| Gene Symbol | TRIM22 |
| Entrez Gene ID | 713814 |
| Full Name | tripartite motif containing 22 |
| Gene Type | protein-coding |
| Organism | Macaca mulatta(Rhesus monkey) |
HOME
ORF » Species Summary » Macaca mulatta » TRIM22 cDNA ORF clone
| Gene Symbol | TRIM22 |
| Entrez Gene ID | 713814 |
| Full Name | tripartite motif containing 22 |
| Gene Type | protein-coding |
| Organism | Macaca mulatta(Rhesus monkey) |
| mRNA | Protein | Name |
|---|---|---|
| XM_015115035.1 | XP_014970521.1 | E3 ubiquitin-protein ligase TRIM22 isoform X1 |
| XM_015115035.2 | XP_014970521.1 | E3 ubiquitin-protein ligase TRIM22 isoform X1 |
| NM_001113359.1 | NP_001106830.1 | E3 ubiquitin-protein ligase TRIM22 |


Genome sequencing and comparison of two nonhuman primate animal models, the cynomolgus and Chinese rhesus macaques.
Yan G, Zhang G, Fang X, Zhang Y, Li C, Ling F, Cooper DN, Li Q, Li Y, van Gool AJ, Du H, Chen J, Chen R, Zhang P, Huang Z, Thompson JR, Meng Y, Bai Y, Wang J, Zhuo M, Wang T, Huang Y, Wei L, Li J, Wang Z, Hu H, Yang P, Le L, Stenson PD, Li B, Liu X, Ball EV, An N, Huang Q, Zhang Y, Fan W, Zhang X, Li Y, Wang W, Katze MG, Su B, Nielsen R, Yang H, Wang J, Wang X, Wang J
Nature biotechnology29(11)1019-23(2011 Oct)
A 5' extended IFN-stimulating response element is crucial for IFN-gamma-induced tripartite motif 22 expression via interaction with IFN regulatory factor-1.
Gao B, Wang Y, Xu W, Duan Z, Xiong S
Journal of immunology (Baltimore, Md. : 1950)185(4)2314-23(2010 Aug)
Different subcellular localisations of TRIM22 suggest species-specific function.
Herr AM, Dressel R, Walter L
Immunogenetics61(4)271-80(2009 Apr)
Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals.
Sawyer SL, Emerman M, Malik HS
PLoS pathogens3(12)e197(2007 Dec)
GeneRIFs: Gene References Into Functions What's a GeneRIF?
Data show that RNA interference abolished IFN-gamma-induced TRIM22 expression, indicating an IRF-1-dependent expression of TRIM22.
Title: A 5' extended IFN-stimulating response element is crucial for IFN-gamma-induced tripartite motif 22 expression via interaction with IFN regulatory factor-1.
Study shows that human and rhesus TRIM22 localise to different subcellular compartments and that this difference can be assigned to the positively selected B30.2 domain.
Title: Different subcellular localisations of TRIM22 suggest species-specific function.
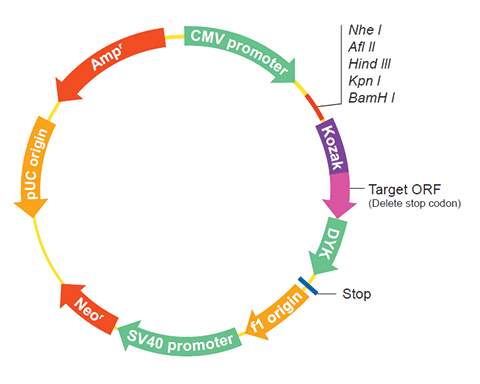
The following TRIM22 gene cDNA ORF clone sequences were retrieved from the NCBI Reference Sequence Database (RefSeq). These sequences represent the protein coding region of the TRIM22 cDNA ORF which is encoded by the open reading frame (ORF) sequence. ORF sequences can be delivered in our standard vector, pcDNA3.1+/C-(K)DYK or the vector of your choice as an expression/transfection-ready ORF clone. Not the clone you want? Click here to find your clone.
| CloneID | OMb02397 | |
| Clone ID Related Accession (Same CDS sequence) | XM_015115035.2 , NM_001113359.1 | |
| Accession Version | NM_001113359.1 Latest version! | Documents for ORF clone product in default vector |
| Sequence Information | ORF Nucleotide Sequence (Length: 1497bp) Protein sequence SNP |
|
| Vector | pcDNA3.1-C-(k)DYK or customized vector |  User Manual User Manual |
| Clone information | Clone Map |  MSDS MSDS |
| Tag on pcDNA3.1+/C-(K)DYK | C terminal DYKDDDDK tags | |
| ORF Insert Method | CloneEZ™ Seamless cloning technology | |
| Insert Structure | linear | |
| Update Date | 2019-04-27 | |
| Organism | Macaca mulatta(Rhesus monkey) | |
| Product | E3 ubiquitin-protein ligase TRIM22 | |
| Comment | Comment: PROVISIONAL REFSEQ: This record has not yet been subject to final NCBI review. The reference sequence was derived from EU124697.1. On or before Jan 10, 2008 this sequence version replaced XM_001110920.1, XM_001111026.1. ##Evidence-Data-START## Transcript exon combination :: EU124697.1 [ECO:0000332] RNAseq introns :: mixed/partial sample support SAMEA4688315, SAMEA4688316 [ECO:0000350] ##Evidence-Data-END## | |
1 | ATGGATTTCT CAGTAAAGGT AGACATAGAG AAGGAGGTGA CCTGCCCCAT CTGCCTGGAG |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
| RefSeq | NP_001106830.1 |
| CDS | 1..1497 |
| Translation |

Target ORF information:
Target ORF information:
|
 NM_001113359.1 NM_001113359.1 |

1 | ATGGATTTCT CAGTAAAGGT AGACATAGAG AAGGAGGTGA CCTGCCCCAT CTGCCTGGAG |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
| CloneID | OMb02397 | |
| Clone ID Related Accession (Same CDS sequence) | XM_015115035.2 , NM_001113359.1 | |
| Accession Version | XM_015115035.2 Latest version! | Documents for ORF clone product in default vector |
| Sequence Information | ORF Nucleotide Sequence (Length: 1497bp) Protein sequence SNP |
|
| Vector | pcDNA3.1-C-(k)DYK or customized vector |  User Manual User Manual |
| Clone information | Clone Map |  MSDS MSDS |
| Tag on pcDNA3.1+/C-(K)DYK | C terminal DYKDDDDK tags | |
| ORF Insert Method | CloneEZ™ Seamless cloning technology | |
| Insert Structure | linear | |
| Update Date | 2019-04-25 | |
| Organism | Macaca mulatta(Rhesus monkey) | |
| Product | E3 ubiquitin-protein ligase TRIM22 isoform X1 | |
| Comment | Comment: MODEL REFSEQ: This record is predicted by automated computational analysis. This record is derived from a genomic sequence (NC_041767.1) annotated using gene prediction method: Gnomon, supported by mRNA and EST evidence. Also see: Documentation of NCBI's Annotation Process On Apr 23, 2019 this sequence version replaced XM_015115035.1. ##Genome-Annotation-Data-START## Annotation Provider :: NCBI Annotation Status :: Full annotation Annotation Name :: Macaca mulatta Annotation Release 103 Annotation Version :: 103 Annotation Pipeline :: NCBI eukaryotic genome annotation pipeline Annotation Software Version :: 8.2 Annotation Method :: Best-placed RefSeq; Gnomon Features Annotated :: Gene; mRNA; CDS; ncRNA ##Genome-Annotation-Data-END## | |
1 | ATGGATTTCT CAGTAAAGGT AGACATAGAG AAGGAGGTGA CCTGCCCCAT CTGCCTGGAG |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
| RefSeq | XP_014970521.1 |
| CDS | 314..1810 |
| Translation |

Target ORF information:
Target ORF information:
|
 XM_015115035.2 XM_015115035.2 |

1 | ATGGATTTCT CAGTAAAGGT AGACATAGAG AAGGAGGTGA CCTGCCCCAT CTGCCTGGAG |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
| CloneID | OMb47382 | |
| Clone ID Related Accession (Same CDS sequence) | XM_015115035.1 | |
| Accession Version | XM_015115035.1 Latest version! | Documents for ORF clone product in default vector |
| Sequence Information | ORF Nucleotide Sequence (Length: 1497bp) Protein sequence SNP |
|
| Vector | pcDNA3.1-C-(k)DYK or customized vector |  User Manual User Manual |
| Clone information | Clone Map |  MSDS MSDS |
| Tag on pcDNA3.1+/C-(K)DYK | C terminal DYKDDDDK tags | |
| ORF Insert Method | CloneEZ™ Seamless cloning technology | |
| Insert Structure | linear | |
| Update Date | 2015-12-20 | |
| Organism | Macaca mulatta(Rhesus monkey) | |
| Product | E3 ubiquitin-protein ligase TRIM22 isoform X1 | |
| Comment | Comment: MODEL REFSEQ: This record is predicted by automated computational analysis. This record is derived from a genomic sequence (NW_014805464.1) annotated using gene prediction method: Gnomon, supported by mRNA and EST evidence. Also see: Documentation of NCBI's Annotation Process ##Genome-Annotation-Data-START## Annotation Provider :: NCBI Annotation Status :: Full annotation Annotation Version :: Macaca mulatta Annotation Release 102 Annotation Pipeline :: NCBI eukaryotic genome annotation pipeline Annotation Software Version :: 6.5 Annotation Method :: Best-placed RefSeq; Gnomon Features Annotated :: Gene; mRNA; CDS; ncRNA ##Genome-Annotation-Data-END## | |
1 | ATGGATTTCT CAGTAAAGGT AGACATAGAG AAGGAGGTGA CCTGCCCCAT CTGCCTGGAG |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
| RefSeq | XP_014970521.1 |
| CDS | 314..1810 |
| Translation |

Target ORF information:
Target ORF information:
|
 XM_015115035.1 XM_015115035.1 |

1 | ATGGATTTCT CAGTAAAGGT AGACATAGAG AAGGAGGTGA CCTGCCCCAT CTGCCTGGAG |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
Genome sequencing and comparison of two nonhuman primate animal models, the cynomolgus and Chinese rhesus macaques. |
A 5' extended IFN-stimulating response element is crucial for IFN-gamma-induced tripartite motif 22 expression via interaction with IFN regulatory factor-1. |
Different subcellular localisations of TRIM22 suggest species-specific function. |
Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals. |