| Gene Symbol | SLC25A15 |
| Entrez Gene ID | 697575 |
| Full Name | solute carrier family 25 member 15 |
| Gene Type | protein-coding |
| Organism | Macaca mulatta(Rhesus monkey) |
HOME
ORF » Species Summary » Macaca mulatta » SLC25A15 cDNA ORF clone
| Gene Symbol | SLC25A15 |
| Entrez Gene ID | 697575 |
| Full Name | solute carrier family 25 member 15 |
| Gene Type | protein-coding |
| Organism | Macaca mulatta(Rhesus monkey) |
| mRNA | Protein | Name |
|---|---|---|
| XM_001088596.3 | XP_001088596.1 | mitochondrial ornithine transporter 1 isoform X1 |
| XM_015120965.1 | XP_014976451.1 | mitochondrial ornithine transporter 1 isoform X2 |

| Pan troglodytes (chimpanzee) | SLC25A15 | XP_003954299.1 |
| SLC25A15 | XP_001088596.1 | |
| Gallus gallus (chicken) | SLC25A15 | NP_001008442.1 |
| Danio rerio (zebrafish) | LOC565335 | NP_001121816.1 |
| Canis lupus familiaris (dog) | SLC25A15 | XP_543118.1 |
| Mus musculus (house mouse) | Slc25a15 | NP_851842.1 |
| Drosophila melanogaster (fruit fly) | CG1628 | NP_001259423.1 |
| Xenopus tropicalis (tropical clawed frog) | slc25a15 | NP_001090866.1 |
| Rattus norvegicus (Norway rat) | Slc25a15 | NP_001041345.1 |
| Danio rerio (zebrafish) | slc25a15a | NP_001074107.1 |
| Homo sapiens (human) | SLC25A15 | NP_055067.1 |
| Bos taurus (cattle) | SLC25A15 | NP_001039791.1 |
| Caenorhabditis elegans (roundworm) | T10F2.2 | NP_498094.1 |

A new rhesus macaque assembly and annotation for next-generation sequencing analyses.
Zimin AV, Cornish AS, Maudhoo MD, Gibbs RM, Zhang X, Pandey S, Meehan DT, Wipfler K, Bosinger SE, Johnson ZP, Tharp GK, Mar?ais G, Roberts M, Ferguson B, Fox HS, Treangen T, Salzberg SL, Yorke JA, Norgren RB Jr
Biology direct9(1)20(2014 Oct)
GeneRIFs: Gene References Into Functions What's a GeneRIF?
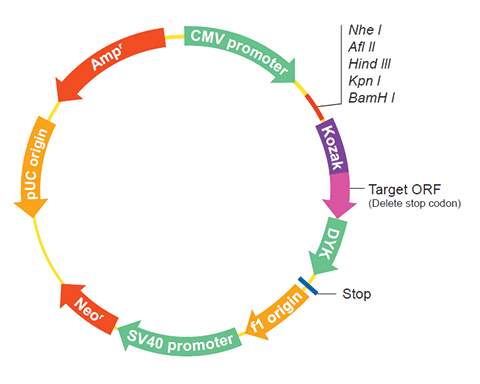
The following SLC25A15 gene cDNA ORF clone sequences were retrieved from the NCBI Reference Sequence Database (RefSeq). These sequences represent the protein coding region of the SLC25A15 cDNA ORF which is encoded by the open reading frame (ORF) sequence. ORF sequences can be delivered in our standard vector, pcDNA3.1+/C-(K)DYK or the vector of your choice as an expression/transfection-ready ORF clone. Not the clone you want? Click here to find your clone.
| CloneID | OMb10995 | |
| Clone ID Related Accession (Same CDS sequence) | XM_001088596.3 | |
| Accession Version | XM_001088596.3 Latest version! | Documents for ORF clone product in default vector |
| Sequence Information | ORF Nucleotide Sequence (Length: 906bp) Protein sequence SNP |
|
| Vector | pcDNA3.1-C-(k)DYK or customized vector |  User Manual User Manual |
| Clone information | Clone Map |  MSDS MSDS |
| Tag on pcDNA3.1+/C-(K)DYK | C terminal DYKDDDDK tags | |
| ORF Insert Method | CloneEZ™ Seamless cloning technology | |
| Insert Structure | linear | |
| Update Date | 2015-12-20 | |
| Organism | Macaca mulatta(Rhesus monkey) | |
| Product | mitochondrial ornithine transporter 1 isoform X1 | |
| Comment | Comment: MODEL REFSEQ: This record is predicted by automated computational analysis. This record is derived from a genomic sequence (NW_014805602.1) and transcript sequence (JV635899.1) annotated using gene prediction method: Gnomon, supported by EST evidence. Also see: Documentation of NCBI's Annotation Process On Dec 21, 2015 this sequence version replaced XM_001088596.2. ##Genome-Annotation-Data-START## Annotation Provider :: NCBI Annotation Status :: Full annotation Annotation Version :: Macaca mulatta Annotation Release 102 Annotation Pipeline :: NCBI eukaryotic genome annotation pipeline Annotation Software Version :: 6.5 Annotation Method :: Best-placed RefSeq; Gnomon Features Annotated :: Gene; mRNA; CDS; ncRNA ##Genome-Annotation-Data-END## ##RefSeq-Attributes-START## assembly gap :: added 116 transcript bases to patch genome assembly gap ##RefSeq-Attributes-END## | |
1 | ATGAAGTCCA ATCCGGCTAT CCAGGCTGCT ATTGACCTCA CGGCGGGGGC TGCAGGGGGC |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
| RefSeq | XP_001088596.1 |
| CDS | 186..1091 |
| Translation |

Target ORF information:
Target ORF information:
|
 XM_001088596.3 XM_001088596.3 |

1 | ATGAAGTCCA ATCCGGCTAT CCAGGCTGCT ATTGACCTCA CGGCGGGGGC TGCAGGGGGC |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
| CloneID | OMb51611 | |
| Clone ID Related Accession (Same CDS sequence) | XM_015120965.1 | |
| Accession Version | XM_015120965.1 Latest version! | Documents for ORF clone product in default vector |
| Sequence Information | ORF Nucleotide Sequence (Length: 807bp) Protein sequence SNP |
|
| Vector | pcDNA3.1-C-(k)DYK or customized vector |  User Manual User Manual |
| Clone information | Clone Map |  MSDS MSDS |
| Tag on pcDNA3.1+/C-(K)DYK | C terminal DYKDDDDK tags | |
| ORF Insert Method | CloneEZ™ Seamless cloning technology | |
| Insert Structure | linear | |
| Update Date | 2015-12-20 | |
| Organism | Macaca mulatta(Rhesus monkey) | |
| Product | mitochondrial ornithine transporter 1 isoform X2 | |
| Comment | Comment: MODEL REFSEQ: This record is predicted by automated computational analysis. This record is derived from a genomic sequence (NW_014805602.1) and transcript sequence (JV635899.1) annotated using gene prediction method: Gnomon, supported by EST evidence. Also see: Documentation of NCBI's Annotation Process ##Genome-Annotation-Data-START## Annotation Provider :: NCBI Annotation Status :: Full annotation Annotation Version :: Macaca mulatta Annotation Release 102 Annotation Pipeline :: NCBI eukaryotic genome annotation pipeline Annotation Software Version :: 6.5 Annotation Method :: Best-placed RefSeq; Gnomon Features Annotated :: Gene; mRNA; CDS; ncRNA ##Genome-Annotation-Data-END## ##RefSeq-Attributes-START## assembly gap :: added 116 transcript bases to patch genome assembly gap ##RefSeq-Attributes-END## | |
1 | ATGAAGTCCA ATCCGGCTAT CCAGGCTGCT ATTGACCTCA CGGCGGGGGC TGCAGGGGGC |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
| RefSeq | XP_014976451.1 |
| CDS | 186..992 |
| Translation |

Target ORF information:
Target ORF information:
|
 XM_015120965.1 XM_015120965.1 |

1 | ATGAAGTCCA ATCCGGCTAT CCAGGCTGCT ATTGACCTCA CGGCGGGGGC TGCAGGGGGC |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
A new rhesus macaque assembly and annotation for next-generation sequencing analyses. |