| Gene Symbol | HSPA5 |
| Entrez Gene ID | 415113 |
| Full Name | heat shock protein family A (Hsp70) member 5 |
| Synonyms | grp78 |
| Gene Type | protein-coding |
| Organism | Bos taurus(cattle) |
HOME
ORF » Species Summary » Bos taurus » HSPA5 cDNA ORF clone
| Gene Symbol | HSPA5 |
| Entrez Gene ID | 415113 |
| Full Name | heat shock protein family A (Hsp70) member 5 |
| Synonyms | grp78 |
| Gene Type | protein-coding |
| Organism | Bos taurus(cattle) |
| mRNA | Protein | Name |
|---|---|---|
| XM_024998380.1 | XP_024854148.1 | endoplasmic reticulum chaperone BiP isoform X1 |
| NM_001075148.1 | NP_001068616.1 | endoplasmic reticulum chaperone BiP precursor |

| Caenorhabditis elegans (roundworm) | hsp-4 | NP_495536.1 |
| Schizosaccharomyces pombe (fission yeast) | bip1 | NP_593245.1 |
| Arabidopsis thaliana (thale cress) | BIP1 | NP_198206.1 |
| Homo sapiens (human) | HSPA5 | NP_005338.1 |
| Rattus norvegicus (Norway rat) | Hspa5 | NP_037215.1 |
| Danio rerio (zebrafish) | hspa5 | NP_998223.1 |
| Neurospora crassa | NCU03982 | XP_956567.1 |
| Xenopus tropicalis (tropical clawed frog) | LOC100492570 | XP_002941690.1 |
| HSPA5 | XP_001099110.1 | |
| Canis lupus familiaris (dog) | HSPA5 | XP_863385.2 |
| Bos taurus (cattle) | HSPA5 | NP_001068616.1 |
| Anopheles gambiae (African malaria mosquito) | AgaP_AGAP004192 | XP_313085.3 |
| Eremothecium gossypii | AGOS_ACR038W | NP_983441.1 |
| Caenorhabditis elegans (roundworm) | hsp-3 | NP_509019.1 |
| Saccharomyces cerevisiae (baker's yeast) | KAR2 | NP_012500.3 |
| Kluyveromyces lactis | KLLA0D09559g | XP_453488.1 |
| Arabidopsis thaliana (thale cress) | BIP2 | NP_851119.1 |
| Pan troglodytes (chimpanzee) | HSPA5 | XP_520257.2 |
| Mus musculus (house mouse) | Hspa5 | NP_001156906.1 |
| Gallus gallus (chicken) | HSPA5 | NP_990822.1 |
| Drosophila melanogaster (fruit fly) | Hsc70-3 | NP_727564.1 |
| Magnaporthe oryzae (rice blast fungus) | MGG_02503 | XP_003709308.1 |
| Oryza sativa (rice) | Os02g0115900 | NP_001045675.1 |

A whole-genome assembly of the domestic cow, Bos taurus.
Zimin AV, Delcher AL, Florea L, Kelley DR, Schatz MC, Puiu D, Hanrahan F, Pertea G, Van Tassell CP, Sonstegard TS, Mar?ais G, Roberts M, Subramanian P, Yorke JA, Salzberg SL
Genome biology10(4)R42(2009)
AB5 subtilase cytotoxin inactivates the endoplasmic reticulum chaperone BiP.
Paton AW, Beddoe T, Thorpe CM, Whisstock JC, Wilce MC, Rossjohn J, Talbot UM, Paton JC
Nature443(7111)548-52(2006 Oct)
Characterization of 954 bovine full-CDS cDNA sequences.
Harhay GP, Sonstegard TS, Keele JW, Heaton MP, Clawson ML, Snelling WM, Wiedmann RT, Van Tassell CP, Smith TP
BMC genomics6166(2005 Nov)
Localization of the chaperone proteins GRP78 and HSP60 on the luminal surface of bovine oviduct epithelial cells and their association with spermatozoa.
Boilard M, Reyes-Moreno C, Lachance C, Massicotte L, Bailey JL, Sirard MA, Leclerc P
Biology of reproduction71(6)1879-89(2004 Dec)
Monitoring gene expression changes in bovine oviduct epithelial cells during the oestrous cycle.
Bauersachs S, Rehfeld S, Ulbrich SE, Mallok S, Prelle K, Wenigerkind H, Einspanier R, Blum H, Wolf E
Journal of molecular endocrinology32(2)449-66(2004 Apr)
GeneRIFs: Gene References Into Functions What's a GeneRIF?
BiP is a master regulator of endoplasmic reticulum function, and its cleavage by subtilase cytotoxin represents a previously unknown trigger for cell death
Title: AB5 subtilase cytotoxin inactivates the endoplasmic reticulum chaperone BiP.
This paper reports the localization of both GRP78 and HSP60 on the luminal/apical surface of oviduct epithelial cells, their binding to spermatozoa, and the presence of endogenous HSP60 in the sperm midpiece.
Title: Localization of the chaperone proteins GRP78 and HSP60 on the luminal surface of bovine oviduct epithelial cells and their association with spermatozoa.
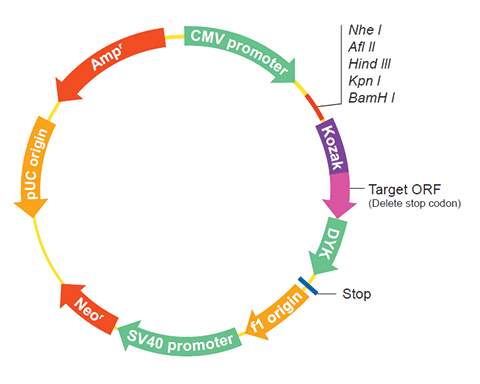
The following HSPA5 gene cDNA ORF clone sequences were retrieved from the NCBI Reference Sequence Database (RefSeq). These sequences represent the protein coding region of the HSPA5 cDNA ORF which is encoded by the open reading frame (ORF) sequence. ORF sequences can be delivered in our standard vector, pcDNA3.1+/C-(K)DYK or the vector of your choice as an expression/transfection-ready ORF clone. Not the clone you want? Click here to find your clone.
| CloneID | OBa42804 | |
| Clone ID Related Accession (Same CDS sequence) | XM_024998380.1 , NM_001075148.1 | |
| Accession Version | NM_001075148.1 Latest version! | Documents for ORF clone product in default vector |
| Sequence Information | ORF Nucleotide Sequence (Length: 1968bp) Protein sequence SNP |
|
| Vector | pcDNA3.1-C-(k)DYK or customized vector |  User Manual User Manual |
| Clone information | Clone Map |  MSDS MSDS |
| Tag on pcDNA3.1+/C-(K)DYK | C terminal DYKDDDDK tags | |
| ORF Insert Method | CloneEZ™ Seamless cloning technology | |
| Insert Structure | linear | |
| Update Date | 2019-12-24 | |
| Organism | Bos taurus(cattle) | |
| Product | endoplasmic reticulum chaperone BiP precursor | |
| Comment | Comment: PROVISIONAL REFSEQ: This record has not yet been subject to final NCBI review. The reference sequence was derived from BC119953.1. On Oct 4, 2006 this sequence version replaced XM_867578.1. ##Evidence-Data-START## Transcript exon combination :: BC119953.1 [ECO:0000332] RNAseq introns :: single sample supports all introns SAMN03145419, SAMN03145442 [ECO:0000348] ##Evidence-Data-END## | |
1 | ATGAAGCTGT CCCTGGTGGC TGCGGTGCTG CTGCTGCTGC TTGGCACGGC ACGCGCGGAG |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
| RefSeq | NP_001068616.1 |
| CDS | 122..2089 |
| Translation |

Target ORF information:
Target ORF information:
|
 NM_001075148.1 NM_001075148.1 |

1 | ATGAAGCTGT CCCTGGTGGC TGCGGTGCTG CTGCTGCTGC TTGGCACGGC ACGCGCGGAG |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
| CloneID | OBa42804 | |
| Clone ID Related Accession (Same CDS sequence) | XM_024998380.1 , NM_001075148.1 | |
| Accession Version | XM_024998380.1 Latest version! | Documents for ORF clone product in default vector |
| Sequence Information | ORF Nucleotide Sequence (Length: 1968bp) Protein sequence SNP |
|
| Vector | pcDNA3.1-C-(k)DYK or customized vector |  User Manual User Manual |
| Clone information | Clone Map |  MSDS MSDS |
| Tag on pcDNA3.1+/C-(K)DYK | C terminal DYKDDDDK tags | |
| ORF Insert Method | CloneEZ™ Seamless cloning technology | |
| Insert Structure | linear | |
| Update Date | 2018-05-06 | |
| Organism | Bos taurus(cattle) | |
| Product | endoplasmic reticulum chaperone BiP isoform X1 | |
| Comment | Comment: MODEL REFSEQ: This record is predicted by automated computational analysis. This record is derived from a genomic sequence (NC_037338.1) annotated using gene prediction method: Gnomon, supported by mRNA and EST evidence. Also see: Documentation of NCBI's Annotation Process ##Genome-Annotation-Data-START## Annotation Provider :: NCBI Annotation Status :: Full annotation Annotation Name :: Bos taurus Annotation Release 106 Annotation Version :: 106 Annotation Pipeline :: NCBI eukaryotic genome annotation pipeline Annotation Software Version :: 8.0 Annotation Method :: Best-placed RefSeq; Gnomon Features Annotated :: Gene; mRNA; CDS; ncRNA ##Genome-Annotation-Data-END## | |
1 | ATGAAGCTGT CCCTGGTGGC TGCGGTGCTG CTGCTGCTGC TTGGCACGGC ACGCGCGGAG |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
| RefSeq | XP_024854148.1 |
| CDS | 259..2226 |
| Translation |

Target ORF information:
Target ORF information:
|
 XM_024998380.1 XM_024998380.1 |

1 | ATGAAGCTGT CCCTGGTGGC TGCGGTGCTG CTGCTGCTGC TTGGCACGGC ACGCGCGGAG |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
A whole-genome assembly of the domestic cow, Bos taurus. |
AB5 subtilase cytotoxin inactivates the endoplasmic reticulum chaperone BiP. |
Characterization of 954 bovine full-CDS cDNA sequences. |
Localization of the chaperone proteins GRP78 and HSP60 on the luminal surface of bovine oviduct epithelial cells and their association with spermatozoa. |
Monitoring gene expression changes in bovine oviduct epithelial cells during the oestrous cycle. |