| Gene Symbol | SBE2.2 |
| Entrez Gene ID | 831769 |
| Full Name | starch branching enzyme 2.2 |
| Synonyms | F17C15.70,F17C15_70,starch branching enzyme 2.2 |
| Gene Type | protein-coding |
| Organism | Arabidopsis thaliana(thale cress) |
HOME
ORF » Species Summary » Arabidopsis thaliana » SBE2.2 cDNA ORF clone
| Gene Symbol | SBE2.2 |
| Entrez Gene ID | 831769 |
| Full Name | starch branching enzyme 2.2 |
| Synonyms | F17C15.70,F17C15_70,starch branching enzyme 2.2 |
| Gene Type | protein-coding |
| Organism | Arabidopsis thaliana(thale cress) |
| mRNA | Protein | Name |
|---|---|---|
| NM_120446.4 | NP_195985.3 | starch branching enzyme 2.2 |

| Gallus gallus (chicken) | GBE1 | XP_425536.4 |
| Danio rerio (zebrafish) | si:ch211-213e17.1 | XP_002663606.3 |
| Canis lupus familiaris (dog) | LOC478380 | XP_535555.3 |
| Rattus norvegicus (Norway rat) | Gbe1 | NP_001093972.1 |
| Arabidopsis thaliana (thale cress) | SBE2.1 | NP_181180.1 |
| Xenopus tropicalis (tropical clawed frog) | gbe1 | XP_004912135.1 |
| Homo sapiens (human) | GBE1 | NP_000149.3 |
| Magnaporthe oryzae (rice blast fungus) | MGG_03186 | XP_003716803.1 |
| Oryza sativa (rice) | Os02g0528200 | NP_001047009.1 |
| Bos taurus (cattle) | GBE1 | NP_001116201.1 |
| Drosophila melanogaster (fruit fly) | CG33138 | NP_001260941.1 |
| Anopheles gambiae (African malaria mosquito) | AgaP_AGAP010428 | XP_311519.3 |
| Kluyveromyces lactis | KLLA0A11176g | XP_451486.1 |
| Eremothecium gossypii | AGOS_AEL044W | NP_984817.1 |
| Neurospora crassa | NCU05429 | XP_963252.2 |
| Arabidopsis thaliana (thale cress) | SBE2.2 | NP_195985.3 |
| Pan troglodytes (chimpanzee) | GBE1 | XP_516593.2 |
| GBE1 | XP_001118968.2 | |
| Mus musculus (house mouse) | Gbe1 | NP_083079.1 |
| Danio rerio (zebrafish) | LOC559212 | XP_687620.6 |
| Caenorhabditis elegans (roundworm) | CELE_T04A8.7 | NP_497961.1 |
| Saccharomyces cerevisiae (baker's yeast) | GLC3 | NP_010905.1 |

Sequence and analysis of chromosome 5 of the plant Arabidopsis thaliana.
Tabata S, Kaneko T, Nakamura Y, Kotani H, Kato T, Asamizu E, Miyajima N, Sasamoto S, Kimura T, Hosouchi T, Kawashima K, Kohara M, Matsumoto M, Matsuno A, Muraki A, Nakayama S, Nakazaki N, Naruo K, Okumura S, Shinpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Sato S, de la Bastide M, Huang E, Spiegel L, Gnoj L, O'Shaughnessy A, Preston R, Habermann K, Murray J, Johnson D, Rohlfing T, Nelson J, Stoneking T, Pepin K, Spieth J, Sekhon M, Armstrong J, Becker M, Belter E, Cordum H, Cordes M, Courtney L, Courtney W, Dante M, Du H, Edwards J, Fryman J, Haakensen B, Lamar E, Latreille P, Leonard S, Meyer R, Mulvaney E, Ozersky P, Riley A, Strowmatt C, Wagner-McPherson C, Wollam A, Yoakum M, Bell M, Dedhia N, Parnell L, Shah R, Rodriguez M, See LH, Vil D, Baker J, Kirchoff K, Toth K, King L, Bahret A, Miller B, Marra M, Martienssen R, McCombie WR, Wilson RK, Murphy G, Bancroft I, Volckaert G, Wambutt R, D?sterh?ft A, Stiekema W, Pohl T, Entian KD, Terryn N, Hartley N, Bent E, Johnson S, Langham SA, McCullagh B, Robben J, Grymonprez B, Zimmermann W, Ramsperger U, Wedler H, Balke K, Wedler E, Peters S, van Staveren M, Dirkse W, Mooijman P, Lankhorst RK, Weitzenegger T, Bothe G, Rose M, Hauf J, Berneiser S, Hempel S, Feldpausch M, Lamberth S, Villarroel R, Gielen J, Ardiles W, Bents O, Lemcke K, Kolesov G, Mayer K, Rudd S, Schoof H, Schueller C, Zaccaria P, Mewes HW, Bevan M, Fransz P, , ,
Nature408(6814)823-6(2000 Dec)
GeneRIFs: Gene References Into Functions What's a GeneRIF?
This work describes the enzymatic parameters of Arabidopsis BE2.2. It reveals for the first time conformational changes for a branching enzyme, leading to a positive cooperative binding process of this enzyme.
Title: Biochemical characterization of Arabidopsis thaliana starch branching enzyme 2.2 reveals an enzymatic positive cooperativity.
The cooperative action of starch synthase and branching enzyme isoforms results in glucans with polymodal chain length distribution similar to amylopectin.
Title: Analysis of the functional interaction of Arabidopsis starch synthase and branching enzyme isoforms reveals that the cooperative action of SSI and BEs results in glucans with polymodal chain length distribution similar to amylopectin.
The following SBE2.2 gene cDNA ORF clone sequences were retrieved from the NCBI Reference Sequence Database (RefSeq). These sequences represent the protein coding region of the SBE2.2 cDNA ORF which is encoded by the open reading frame (ORF) sequence. ORF sequences can be delivered in our standard vector, pcDNA3.1+/C-(K)DYK or the vector of your choice as an expression/transfection-ready ORF clone. Not the clone you want? Click here to find your clone.
| CloneID | OAb18832 | |
| Clone ID Related Accession (Same CDS sequence) | NM_120446.4 | |
| Accession Version | NM_120446.4 Latest version! | Documents for ORF clone product in default vector |
| Sequence Information | ORF Nucleotide Sequence (Length: 2418bp) Protein sequence SNP |
|
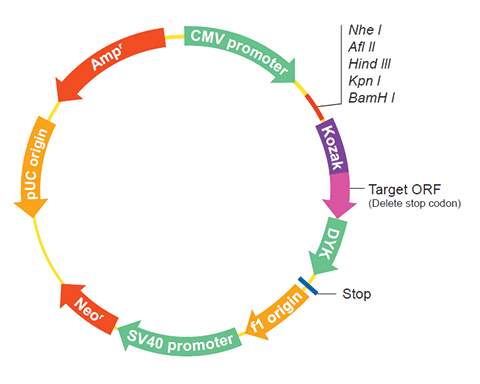
| Vector | pcDNA3.1-C-(k)DYK or customized vector |  User Manual User Manual |
| Clone information | Clone Map |  MSDS MSDS |
| Tag on pcDNA3.1+/C-(K)DYK | C terminal DYKDDDDK tags | |
| ORF Insert Method | CloneEZ™ Seamless cloning technology | |
| Insert Structure | linear | |
| Update Date | 2019-02-13 | |
| Organism | Arabidopsis thaliana(thale cress) | |
| Product | starch branching enzyme 2.2 | |
| Comment | Comment: REVIEWED REFSEQ: This record has been curated by TAIR and Araport. This record is derived from an annotated genomic sequence (NC_003076). On Sep 12, 2016 this sequence version replaced NM_120446.3. | |
1 | ATGGTGGTGA TTCACGGAGT GTCTCTTACT CCACGCTTCA CTCTTCCTTC TCGACCTCTC |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
| RefSeq | NP_195985.3 |
| CDS | 163..2580 |
| Translation |

Target ORF information:
Target ORF information:
|
 NM_120446.4 NM_120446.4 |

1 | ATGGTGGTGA TTCACGGAGT GTCTCTTACT CCACGCTTCA CTCTTCCTTC TCGACCTCTC |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
Sequence and analysis of chromosome 5 of the plant Arabidopsis thaliana. |