| Gene Symbol | SPO11 |
| Entrez Gene ID | 856364 |
| Full Name | Spo11p |
| Gene Type | protein-coding |
| Organism | Saccharomyces cerevisiae S288C |
HOME
ORF » Species Summary » Saccharomyces cerevisiae S288C » SPO11 cDNA ORF clone
| Gene Symbol | SPO11 |
| Entrez Gene ID | 856364 |
| Full Name | Spo11p |
| Gene Type | protein-coding |
| Organism | Saccharomyces cerevisiae S288C |
| mRNA | Protein | Name |
|---|---|---|
| NM_001179102.1 | NP_011841.1 | DNA topoisomerase (ATP-hydrolyzing) |

| Eremothecium gossypii | AGOS_ADR025W | NP_984121.1 |
| Kluyveromyces lactis | KLLA0D14575g | XP_453708.1 |
| Saccharomyces cerevisiae (baker's yeast) | SPO11 | NP_011841.1 |

Life with 6000 genes.
Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG
Science (New York, N.Y.)274(5287)546, 563-7(1996 Oct)
Complete nucleotide sequence of Saccharomyces cerevisiae chromosome VIII.
Johnston M, Andrews S, Brinkman R, Cooper J, Ding H, Dover J, Du Z, Favello A, Fulton L, Gattung S
Science (New York, N.Y.)265(5181)2077-82(1994 Sep)
GeneRIFs: Gene References Into Functions What's a GeneRIF?
Data show that increases in meiotic double-strand breaks and crossover frequencies can be programmed to arbitrary genomic sites by fusion of the Spo11 protein to various DNA-recognition domains.
Title: Programming sites of meiotic crossovers using Spo11 fusion proteins.
A tel1Delta mutant has globally increased amounts of Spo11-oligonucleotide complexes and altered Spo11-oligonucleotide lengths, consistent with conserved roles for Tel1 in control of DSB number and processing..We further find that effects of Tel1 are distinct but partially overlapping with previously described contributions of the recombination regulator Cst9 (also known as Zip3).
Title: Numerical and spatial patterning of yeast meiotic DNA breaks by Tel1.
we found a role for the meiotic bouquet in establishing the size dependence of centromere coupling, as abolishing bouquet (using the bouquet-defective spo11 ndj1 mutant) reduces it. Coupling in spo11 ndj1 rather follows telomere clustering preferences. We propose that a chromosome size preference for centromere coupling helps establish efficient homolog recognition.
Title: Multiple Pairwise Analysis of Non-homologous Centromere Coupling Reveals Preferential Chromosome Size-Dependent Interactions and a Role for Bouquet Formation in Establishing the Interaction Pattern.
We examined fine-scale DSB distributions in TF mutant strains by deep sequencing oligonucleotides that remain covalently bound to Spo11 as a byproduct of DSB formation, mapped Bas1 and Ino4 binding sites in meiotic cells
Title: High-Resolution Global Analysis of the Influences of Bas1 and Ino4 Transcription Factors on Meiotic DNA Break Distributions in Saccharomyces cerevisiae.
These findings provide an explanation for the unexpectedly low prototroph levels exhibited by spo11 hypomorphs and have important implications for genetic studies that assume an unbiased recovery of prototrophs
Title: High throughput sequencing reveals alterations in the recombination signatures with diminishing Spo11 activity.
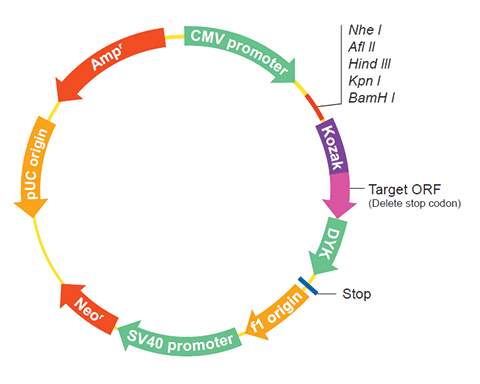
The following SPO11 gene cDNA ORF clone sequences were retrieved from the NCBI Reference Sequence Database (RefSeq). These sequences represent the protein coding region of the SPO11 cDNA ORF which is encoded by the open reading frame (ORF) sequence. ORF sequences can be delivered in our standard vector, pcDNA3.1+/C-(K)DYK or the vector of your choice as an expression/transfection-ready ORF clone. Not the clone you want? Click here to find your clone.
| CloneID | OSi05173 | |
| Clone ID Related Accession (Same CDS sequence) | NM_001179102.1 | |
| Accession Version | NM_001179102.1 Latest version! | Documents for ORF clone product in default vector |
| Sequence Information | ORF Nucleotide Sequence (Length: 1197bp) Protein sequence SNP |
|
| Vector | pcDNA3.1-C-(k)DYK or customized vector |  User Manual User Manual |
| Clone information | Clone Map |  MSDS MSDS |
| Tag on pcDNA3.1+/C-(K)DYK | C terminal DYKDDDDK tags | |
| ORF Insert Method | CloneEZ™ Seamless cloning technology | |
| Insert Structure | linear | |
| Update Date | 2019-10-31 | |
| Organism | Saccharomyces cerevisiae S288C | |
| Product | DNA topoisomerase (ATP-hydrolyzing) | |
| Comment | Comment: REVIEWED REFSEQ: This record has been curated by SGD. This record is derived from an annotated genomic sequence (NC_001140). ##Genome-Annotation-Data-START## Annotation Provider :: SGD Annotation Status :: Full Annotation Annotation Veresion :: R64-2-1 URL :: http://www.yeastgenome.org/ ##Genome-Annotation-Data-END## COMPLETENESS: incomplete on both ends. | |
1 | ATGGCTTTGG AGGGATTGCG GAAAAAATAT AAAACAAGGC AGGAATTGGT CAAAGCACTC |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
| RefSeq | NP_011841.1 |
| CDS | 1..1197 |
| Translation |

Target ORF information:
Target ORF information:
|
 NM_001179102.1 NM_001179102.1 |

1 | ATGGCTTTGG AGGGATTGCG GAAAAAATAT AAAACAAGGC AGGAATTGGT CAAAGCACTC |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
Life with 6000 genes. |
Complete nucleotide sequence of Saccharomyces cerevisiae chromosome VIII. |