| Gene Symbol | grim |
| Entrez Gene ID | 40014 |
| Full Name | CG4345 gene product from transcript CG4345-RA |
| Synonyms | BcDNA:RE28551,CG4345,Dmel\CG4345,GRIM,Grim,gri,grm |
| Gene Type | protein-coding |
| Organism | Drosophila melanogaster(fruit fly) |
| Genome |
HOME
ORF » Species Summary » Drosophila melanogaster » grim cDNA ORF clone
| Gene Symbol | grim |
| Entrez Gene ID | 40014 |
| Full Name | CG4345 gene product from transcript CG4345-RA |
| Synonyms | BcDNA:RE28551,CG4345,Dmel\CG4345,GRIM,Grim,gri,grm |
| Gene Type | protein-coding |
| Organism | Drosophila melanogaster(fruit fly) |
| Genome |
| mRNA | Protein | Name |
|---|---|---|
| NM_079413.3 | NP_524137.2 | grim |


Gene Model Annotations for Drosophila melanogaster: Impact of High-Throughput Data.
Matthews BB, Dos Santos G, Crosby MA, Emmert DB, St Pierre SE, Gramates LS, Zhou P, Schroeder AJ, Falls K, Strelets V, Russo SM, Gelbart WM,
G3 (Bethesda, Md.)5(8)1721-36(2015 Jun)
Gene Model Annotations for Drosophila melanogaster: The Rule-Benders.
Crosby MA, Gramates LS, Dos Santos G, Matthews BB, St Pierre SE, Zhou P, Schroeder AJ, Falls K, Emmert DB, Russo SM, Gelbart WM,
G3 (Bethesda, Md.)5(8)1737-49(2015 Jun)
Sequence finishing and mapping of Drosophila melanogaster heterochromatin.
Hoskins RA, Carlson JW, Kennedy C, Acevedo D, Evans-Holm M, Frise E, Wan KH, Park S, Mendez-Lago M, Rossi F, Villasante A, Dimitri P, Karpen GH, Celniker SE
Science (New York, N.Y.)316(5831)1625-8(2007 Jun)
The Release 5.1 annotation of Drosophila melanogaster heterochromatin.
Smith CD, Shu S, Mungall CJ, Karpen GH
Science (New York, N.Y.)316(5831)1586-91(2007 Jun)
Combined evidence annotation of transposable elements in genome sequences.
Quesneville H, Bergman CM, Andrieu O, Autard D, Nouaud D, Ashburner M, Anxolabehere D
PLoS computational biology1(2)166-75(2005 Jul)
GeneRIFs: Gene References Into Functions What's a GeneRIF?
Dm myb and grim are both involved in regulating PCD in two distinct settings suggests that these two genes may often work together to mediate PCD.
Title: The dREAM/Myb-MuvB complex and Grim are key regulators of the programmed death of neural precursor cells at the Drosophila posterior wing margin.
Study provide evidence that miR-6/11 functions through regulation of the proapoptotic genes, reaper (rpr), head involution defective (hid), grim and sickle (skl).
Title: Overlapping functions of microRNAs in control of apoptosis during Drosophila embryogenesis.
findings demonstrate that neuroblast apoptosis during embryogenesis requires the coordinated expression of the cell death genes grim and reaper, and possibly sickle
Title: Coordinated expression of cell death genes regulates neuroblast apoptosis.
Programmed cell death of microchaete glial cells represents an exceptional opportunity to study the mitochondrial proapoptotic process induced by Grim.
Title: grim promotes programmed cell death of Drosophila microchaete glial cells.
Rpr is able to self-associate and also binds to Hid and Grim.
Title: Drosophila IAP antagonists form multimeric complexes to promote cell death.
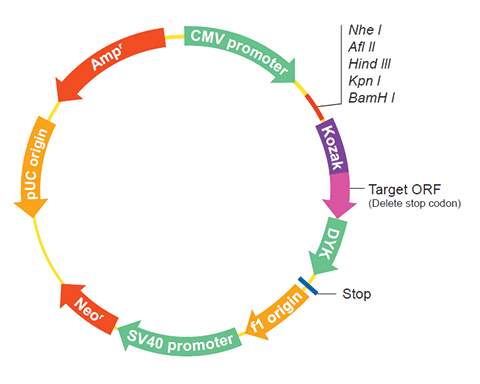
The following grim gene cDNA ORF clone sequences were retrieved from the NCBI Reference Sequence Database (RefSeq). These sequences represent the protein coding region of the grim cDNA ORF which is encoded by the open reading frame (ORF) sequence. ORF sequences can be delivered in our standard vector, pcDNA3.1+/C-(K)DYK or the vector of your choice as an expression/transfection-ready ORF clone. Not the clone you want? Click here to find your clone.
| CloneID | OFa18855 | |
| Clone ID Related Accession (Same CDS sequence) | NM_079413.3 | |
| Accession Version | NM_079413.3 Latest version! | Documents for ORF clone product in default vector |
| Sequence Information | ORF Nucleotide Sequence (Length: 417bp) Protein sequence SNP |
|
| Vector | pcDNA3.1-C-(k)DYK or customized vector |  User Manual User Manual |
| Clone information | Clone Map |  MSDS MSDS |
| Tag on pcDNA3.1+/C-(K)DYK | C terminal DYKDDDDK tags | |
| ORF Insert Method | CloneEZ™ Seamless cloning technology | |
| Insert Structure | linear | |
| Update Date | 2019-04-24 | |
| Organism | Drosophila melanogaster(fruit fly) | |
| Product | grim | |
| Comment | Comment: REVIEWED REFSEQ: This record has been curated by FlyBase. This record is derived from an annotated genomic sequence (NT_037436). On Jan 16, 2013 this sequence version replaced NM_079413.2. ##Genome-Annotation-Data-START## Annotation Provider :: FlyBase Annotation Status :: Full annotation Annotation Version :: Release 6.26 URL :: http://flybase.org ##Genome-Annotation-Data-END## | |
1 | ATGGCCATCG CCTATTTCAT ACCCGACCAG GCCCAATTGT TGGCCAGAAG CTATCAGCAA |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
| RefSeq | NP_524137.2 |
| CDS | 319..735 |
| Translation |

Target ORF information:
Target ORF information:
|
 NM_079413.3 NM_079413.3 |

1 | ATGGCCATCG CCTATTTCAT ACCCGACCAG GCCCAATTGT TGGCCAGAAG CTATCAGCAA |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
Gene Model Annotations for Drosophila melanogaster: Impact of High-Throughput Data. |
Gene Model Annotations for Drosophila melanogaster: The Rule-Benders. |
Sequence finishing and mapping of Drosophila melanogaster heterochromatin. |
The Release 5.1 annotation of Drosophila melanogaster heterochromatin. |
Combined evidence annotation of transposable elements in genome sequences. |