| Gene Symbol | RNR3 |
| Entrez Gene ID | 854744 |
| Full Name | ribonucleotide-diphosphate reductase subunit RNR3 |
| Synonyms | DIN1,RIR3 |
| Gene Type | protein-coding |
| Organism | Saccharomyces cerevisiae S288C |
HOME
ORF » Species Summary » Saccharomyces cerevisiae S288C » RNR3 cDNA ORF clone
| Gene Symbol | RNR3 |
| Entrez Gene ID | 854744 |
| Full Name | ribonucleotide-diphosphate reductase subunit RNR3 |
| Synonyms | DIN1,RIR3 |
| Gene Type | protein-coding |
| Organism | Saccharomyces cerevisiae S288C |
| mRNA | Protein | Name |
|---|---|---|
| NM_001179416.3 | NP_012198.3 | ribonucleotide-diphosphate reductase subunit RNR3 |

| Canis lupus familiaris (dog) | RRM1 | XP_534027.2 |
| Mus musculus (house mouse) | Rrm1 | NP_033129.2 |
| Gallus gallus (chicken) | RRM1 | NP_001026008.1 |
| Saccharomyces cerevisiae (baker's yeast) | RNR3 | NP_012198.3 |
| Eremothecium gossypii | AGOS_ADL057W | NP_984039.2 |
| Schizosaccharomyces pombe (fission yeast) | cdc22 | NP_594491.1 |
| Pan troglodytes (chimpanzee) | RRM1 | XP_001160029.1 |
| Bos taurus (cattle) | RRM1 | NP_001106767.1 |
| Rattus norvegicus (Norway rat) | Rrm1 | NP_001013254.1 |
| Caenorhabditis elegans (roundworm) | rnr-1 | NP_499039.1 |
| Saccharomyces cerevisiae (baker's yeast) | RNR1 | NP_010993.1 |
| Oryza sativa (rice) | Os06g0168600 | NP_001056926.1 |
| Danio rerio (zebrafish) | rrm1 | NP_571530.1 |
| Magnaporthe oryzae (rice blast fungus) | MGG_07000 | XP_003709756.1 |
| Neurospora crassa | NCU03539 | XP_956818.1 |
| Xenopus tropicalis (tropical clawed frog) | rrm1 | NP_001120566.1 |
| Homo sapiens (human) | RRM1 | NP_001024.1 |
| RRM1 | XP_001113010.1 | |
| Drosophila melanogaster (fruit fly) | RnrL | NP_477027.1 |
| Anopheles gambiae (African malaria mosquito) | AgaP_AGAP010198 | XP_554103.3 |
| Kluyveromyces lactis | KLLA0C07887g | XP_452551.1 |
| Arabidopsis thaliana (thale cress) | RNR1 | NP_179770.1 |

The nucleotide sequence of Saccharomyces cerevisiae chromosome IX.
Churcher C, Bowman S, Badcock K, Bankier A, Brown D, Chillingworth T, Connor R, Devlin K, Gentles S, Hamlin N, Harris D, Horsnell T, Hunt S, Jagels K, Jones M, Lye G, Moule S, Odell C, Pearson D, Rajandream M, Rice P, Rowley N, Skelton J, Smith V, Barrell B
Nature387(6632 Suppl)84-7(1997 May)
Life with 6000 genes.
Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG
Science (New York, N.Y.)274(5287)546, 563-7(1996 Oct)
GeneRIFs: Gene References Into Functions What's a GeneRIF?
The gene encoding a large subunit of ribonucleotide reductase, RNR3, is regulated by ISW2 and SWI/SNF, complexes that repress and activate transcription, respectively. Here, we studied the functional interactions of these two complexes at RNR3.
Title: A novel mechanism of antagonism between ATP-dependent chromatin remodeling complexes regulates RNR3 expression.
RNR3 is dependent upon both TFIID and SAGA, two complexes that deliver TATA-binding protein (TBP) to promoters.
Title: Dissection of coactivator requirement at RNR3 reveals unexpected contributions from TFIID and SAGA.
Recruitment of Rap1 to the RNR3 gene is dependent on activation of the DNA damage checkpoint and chromatin remodelling by SWI/SNF.
Title: Yeast Rap1 contributes to genomic integrity by activating DNA damage repair genes.
nucleosome placement plays a dominant role in repression and that the ability of the core promoter to position a nucleosome is a major determinant in TAF(II) dependency of genes in vivo
Title: Exposing the core promoter is sufficient to activate transcription and alter coactivator requirement at RNR3.
Two histone deacetylases, RPD3 and HOS2, are required for the activation of DNA damage-inducible genes RNR3 and HUG1.
Title: Histone deacetylases RPD3 and HOS2 regulate the transcriptional activation of DNA damage-inducible genes.
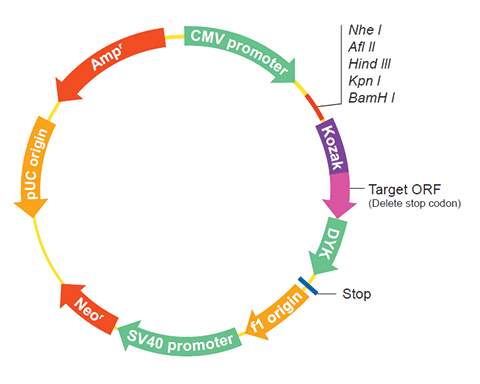
The following RNR3 gene cDNA ORF clone sequences were retrieved from the NCBI Reference Sequence Database (RefSeq). These sequences represent the protein coding region of the RNR3 cDNA ORF which is encoded by the open reading frame (ORF) sequence. ORF sequences can be delivered in our standard vector, pcDNA3.1+/C-(K)DYK or the vector of your choice as an expression/transfection-ready ORF clone. Not the clone you want? Click here to find your clone.
| CloneID | OSi03772 | |
| Clone ID Related Accession (Same CDS sequence) | NM_001179416.3 | |
| Accession Version | NM_001179416.3 Latest version! | Documents for ORF clone product in default vector |
| Sequence Information | ORF Nucleotide Sequence (Length: 2610bp) Protein sequence SNP |
|
| Vector | pcDNA3.1-C-(k)DYK or customized vector |  User Manual User Manual |
| Clone information | Clone Map |  MSDS MSDS |
| Tag on pcDNA3.1+/C-(K)DYK | C terminal DYKDDDDK tags | |
| ORF Insert Method | CloneEZ™ Seamless cloning technology | |
| Insert Structure | linear | |
| Update Date | 2019-10-31 | |
| Organism | Saccharomyces cerevisiae S288C | |
| Product | ribonucleotide-diphosphate reductase subunit RNR3 | |
| Comment | Comment: REVIEWED REFSEQ: This record has been curated by SGD. This record is derived from an annotated genomic sequence (NC_001141). On Jul 30, 2012 this sequence version replaced NM_001179416.2. ##Genome-Annotation-Data-START## Annotation Provider :: SGD Annotation Status :: Full Annotation Annotation Version :: R64-2-1 URL :: http://www.yeastgenome.org/ ##Genome-Annotation-Data-END## COMPLETENESS: incomplete on both ends. | |
1 | ATGTACGTTA TTAAAAGAGA CGGCCGCAAA GAGCCCGTTC AATTCGATAA AATTACCTCC |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
| RefSeq | NP_012198.3 |
| CDS | 1..2610 |
| Translation |

Target ORF information:
Target ORF information:
|
 NM_001179416.3 NM_001179416.3 |

1 | ATGTACGTTA TTAAAAGAGA CGGCCGCAAA GAGCCCGTTC AATTCGATAA AATTACCTCC |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
The nucleotide sequence of Saccharomyces cerevisiae chromosome IX. |
Life with 6000 genes. |